- Home
- Member Resources
- Articles
- Testing of Specimens From Patients With Advanced Colorectal Carcinoma
The advent of new molecularly targeted treatment strategies for patients with advanced colorectal cancer (CRC) places anatomic and molecular pathologists as key players in the delivery of care for this large patient population. Advanced CRC encompasses patients presenting with Stage IV cancer, or those who experience a cancer recurrence. An ever-growing body of literature clearly demonstrates defined opportunities to significantly impact these patients’ lives with the use of targeted medications, the indications of which are derived from molecular testing. In CRC patients, these therapies are currently recommended only after failure of standard first line therapies.
The pathologist’s role in this context is multi-dimensional and incorporates construction of a mechanism to quickly process test requests; ensure that appropriate and adequate specimens are submitted for testing; that specimens are reported in a timely manner; that the relevant information is easily discernible on the pathology report; and that the reports are made readily available to treating clinicians.
With the incorporation of molecular testing as a standard of care, optimization of reflex testing protocols greatly facilitates receipt of timely results to clinical care teams. Reflex testing of all CRCs at first diagnosis to identify patients with a likelihood of Lynch Syndrome is now standard in most US hospitals as well as extended RAS testing in all cases of advanced CRC.
Mismatch Repair [MMR]
The majority of US institutions use a four antibody immunohistochemistry panel to detect loss of protein expression of the canonical mismatch repair genes – MLH1, PMS2, MSH2 and MSH6. EPCAM germline mutations enter the differential via the mutation’s effect on loss of MSH2 protein expression caused by a secondary epigenetic silencing of MSH2.1 PCR-based microsatellite instability testing which detects abnormal patterns of microsatellite accumulations in cancers with MMR deficiency [dMMR] is an equally valid method of dMMR detection. These are reported as microsatellite stable [MSS], microsatellite instability – low [MSI-L] and microsatellite instability- high [MSI-H] based on the modified Bethesda criteria.2 Approximately 15% of all CRCs are dMMR – with about one third of these being derived from Lynch Syndrome. Identification of dMMR status is also associated with improved prognosis for whom 5-FU based therapies in patients with low stage CRCs is contraindicated.
DNA mutations accumulate in large numbers in dMMR CRCs - secondary to an inability to repair spontaneous foci of DNA damage – esp. in regions of repetitive nucleotide sequences. This high mutation load leads to production of large numbers of Mutation Associated Neoantigens [MANAs] on the surface of affected cancer cells. MANAs are prone to elicit a brisk T-cell mediated host anti-tumor immune response.
This host-mediated immune response likely plays a key role in explaining why patients with dMMR CRCs have a better overall survival than those with proficient MMR [pMMR] CRCs.
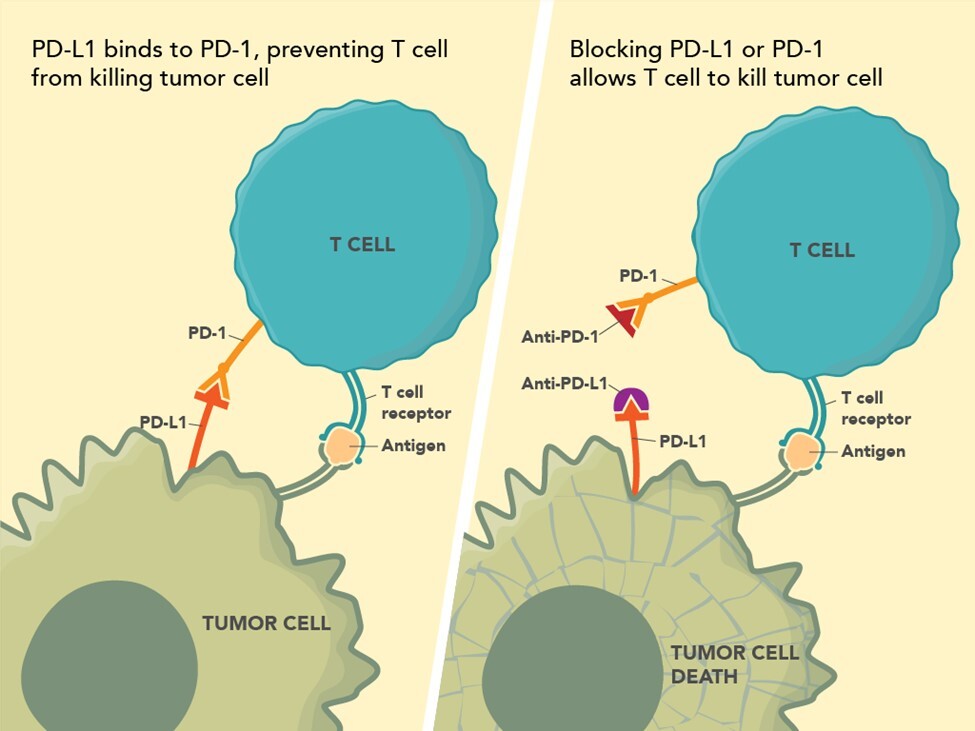
The recognition that innate host responses to tumors are impeded by tumor generated immune suppression pathways has led to remarkable breakthroughs in cancer therapy with the advent of check point inhibitors. Briefly, there are two main pathways currently recognized as important for cancer therapy – the Program Death-1 [PD-1] and the CTLA-4 pathway3 (see Figure 1; Source: US Food and Drug Administration). The Program Death-1 [PD-1] pathway is a naturally occurring T-cell mediated system that prevents the occurrence of autoimmune disease by inhibiting immune responses to normal cells. The PD-1 receptor is on the surface of infiltrating T-cells. PD-L1, the ligand for PD-1, is highly expressed in many cancers. This activated pathway is exploited by cancers to evade innate immune surveillance which might otherwise prove lethal to the tumor. Immune checkpoint inhibitors modulate cytotoxic T-lymphocyte responses against cancer by inhibiting their negative regulation. The PD-1 pathway is important in dMMR colon carcinogenesis.
In a landmark study in 2015, treatment with anti-PD-1 antibody in patients with progressive dMMR CRC was found to have an immune related objective response rate and an immune-related progression free survival rate – compared to patients with progressive CRCs which had pMMR4. Though only a relative small percent of advanced CRCs are dMMR, it is important to ensure that CRCs from these patients are tested for MMR status. Two anti-PD-1 drugs, nivolumab and pembrolizumab, are approved by the FDA for treating CRC patients. However, unlike in other cancer systems, immunohistochemistry for PD-1/PD-L1 has no role in identifying CRC patients who would benefit from anti-PD-1 therapy.
Extended RAS Testing
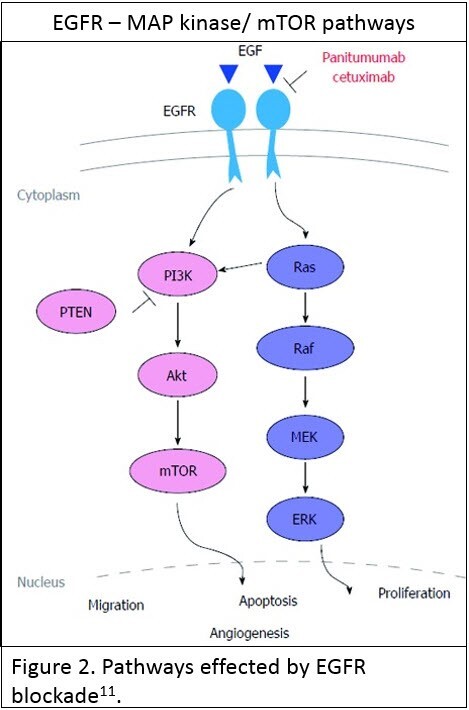
Testing for mutations in the extended RAS family of genes [KRAS and NRAS] is now standard of care before institution of anti-Epidermal Growth Factor Receptor [anti-EGFR] therapy in the presence of advanced CRC. EGFR, which is a cell surface tyrosine kinase, is one of the more important regulators of vital cellular processes such as proliferation and opposing apoptosis, through interaction with multiple pathways – the most important being the RAS-MAP kinase and the AKT-PI3K-mTOR pathways (see Figure 2; reused under license CC BY NC 4.0 ). When stimulated in cancer these pathways have an oncogenic effect facilitating cancer progression.
Inhibition of these pathways by anti-EGFR therapy is only effective if these pathways are primarily stimulated by activation of the EGFR itself. Oncogenic stimulation of the pathways beyond EGFR renders inhibition of the pathway by anti-EGFR ineffective in terms of inhibiting cancer progression.
The RAS family of genes are downstream regulators of EGFR’s effect on the MAP kinase and AKT-PI3K-mTOR pathways. A RAS mutation has an oncogenic stimulatory effect on these pathways such that inhibition of EGFR is ineffective at preventing these oncogenic effects. A KRAS or NRAS mutation is a contraindication for the use of anti-EGFR therapy due to a poor therapeutic response to either of the two FDA approved therapies – panitumumab and cetuximab. Thus testing for the presence of a KRAS or NRAS mutation [Extended RAS] is integral to deciding whether patients may benefit from anti-EGFR therapy.
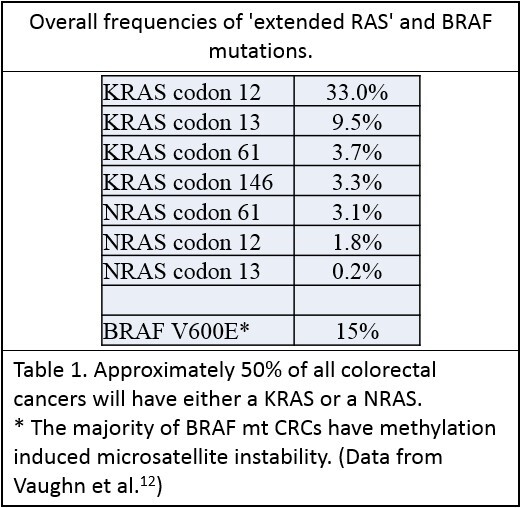
Mutations in KRAS or NRAS genes occur in approximately 50% of CRCs - with the majority occurring in KRAS5. These are single base missense mutations, the majority of which occur in the same codons of each gene, presumably related to similarities between the structures of these genes6. Table 1:The 2017 joint CAP/ASCP/ASCO guidelines recommend that the standard extended RAS panel include testing of KRAS and NRAS codons 12 and 13 of exon 2; 59 and 61 of exon 3; and 117 and 146 of exon 4.7
Treating patients whose CRCs are without KRAS or NRAS mutations with anti-EGFR therapy has been extensively studied. Unfortunately, the overall survival advantage is only of two to three months for this subgroup of patients8.
Tissue for Testing
The 2017 joint CAP/ASCP/ASCO guidelines state that testing for extended RAS mutations is preferably performed on metastatic lesions. Formalin-fixed paraffin embedded tissue is preferred but cytology specimens can be used after validation of the testing methodology with cytology specimens has been performed. In the event of not having specimens from a metastatic lesion, tissue from the primary tumor is commonly used.7 This reflects the greater stability of driver gene mutations in CRC metastases when compared to their respective primaries as opposed to other cancer types.
BRAF V600E
The 2017 joint society guidelines recommended testing for BRAF V600E, as BRAF mutation is also a recognized significant prognostic biomarker of poor outcome in mismatch repair pMMR CRC patients. This latter group accounts for approximately 5% of all CRCs.7
Testing Platforms
Next generation sequencing increasingly has become the standard platform for extended RAS testing due to its versatility to test for multiple gene mutations in a cost effective manner using relatively small amounts of tissue.
Emerging Concepts
Mismatch Repair Testing
Recent reports describe the reliable assessment of MMR status using cancer type specific microsatellite panels using NGS platforms. There are multiple advantages to this approach in many clinical circumstances, and it is likely this methodology will become widely used in the near future.9
Resistance to Target Therapies
Resistance to targeted therapies has become a major problem in modern oncology. Development of anti-EGFR therapy resistance in the setting of initially RAS wild type cancers is virtually inevitable. The most likely mechanism is that of a proliferation of pre-existing cancer sub clones with RAS mutations which become dominant after anti-EGFR therapy decreases the population of RAS wild type cancer cells.
Oncogenic stimuli of the MAP kinase/AKT-mTOR pathways may also have a significant impact on the response to anti-EGFR therapy. Though not currently recommended for routine clinical testing, it is likely that extended studies of these pathways will better predict patient responses. Such candidates, which are the currently the subject of intensive study include various anomalies related to EGFR, BRAF, PTEN, and PIK3CA in particular. Treatment recommendations based on molecular findings outside of current guidelines should be considered in the context of clinical trials. Importantly, the joint societies guidelines stated that the presence of a BRAF mutation, regarded as a MAP kinase oncogenic signal, in a RAS wild type CRC is not regarded as a contraindication to anti-EGFR therapy.7
The continued development of liquid biopsy technologies will likely enter clinical care through monitoring the development of resistance.10 Newer targeted therapeutics and an improved understanding of the mechanisms of CRC progression, will drive an expansion of complex testing with multi-layered data integration, advancing this field in the near future and making a significant impact in improving the survival of patients with advanced CRC.
References
1. Roth RM, Hampel H, Arnold CA, Yearsley MM, Marsh WL, Frankel WL. A modified Lynch syndrome screening algorithm in colon cancer: BRAF immunohistochemistry is efficacious and cost beneficial. Am J Clin Pathol. 2015;143(3):336-343
2. Gologan A, Krasinskas A, Hunt J, Thull DL, Farkas L, Sepulveda AR. Performance of the revised Bethesda guidelines for identification of colorectal carcinomas with a high level of microsatellite instability. Arch Pathol Lab Med. 2005;129(11):1390-1397.
3. Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359(6382):1350-1355.
4. Le DT, Uram JN, Wang H, , et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372(26):2509-2520.
5. Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13(11):828-851.
6. Saeed O, Lopez-Beltran A, Fisher KW, Scarpelli M, Montironi R, Cimadamore A, et al. RAS genes in colorectal carcinoma: pathogenesis, testing guidelines and treatment implications. J Clin Pathol. 2019;72(2):135-139.
7. Sepulveda AR, Hamilton SR, Allegra CJ, et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and American Society of Clinical Oncology. Arch Pathol Lab Med. 2017;141(5):625-657.
8. Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369(11):1023-1034.
9. Hampel H, Pearlman R, Beightol M, et al. Assessment of Tumor Sequencing as a Replacement for Lynch Syndrome Screening and Current Molecular Tests for Patients With Colorectal Cancer. JAMA Oncol. 2018;4(6):806-813.
10. Garcia-Foncillas J, Tabernero J, Elez E, et al. Prospective multicenter real-world RAS mutation comparison between OncoBEAM-based liquid biopsy and tissue analysis in metastatic colorectal cancer. Br J Cancer. 2018;119(12):1464-1470.
11. Jeong WJ, Cha PH, Choi KY. Strategies to overcome resistance to epidermal growth factor receptor monoclonal antibody therapy in metastatic colorectal cancer. World J Gastroenterol. 2014;20(29):9862-9871. Reused under license CC BY NC 4.0.
12. Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes, Chromosomes Cancer. 2011;50(5):307-312.

Joseph E. Willis, MD, FCAP, is a gastrointestinal pathologist in Cleveland, Ohio, and is a professor and vice chair for Translational Research in the Department of Pathology at Case Western Reserve University and University Hospitals Cleveland Medical Center. He received his medical degree from University College Cork Faculty of Medicine and is a fellow of the Royal College of Surgeons in Ireland.