- Home
- Member Resources
- Articles
- Optical Genome Mapping: A ‘Tool’ with Significant Potential from Discovery to Diagnostics
Driven by technological advances, the quest for precision medicine has ushered the ‘omics’ era into clinical practice.1,2 Increasingly, comprehensive sequencing approaches are being deployed to support diagnoses and/or for the detection of actionable findings.3,4 In most clinical diagnostic laboratories, an intriguing dichotomy has emerged in which innovative genomic studies have largely centered on detecting/excluding single nucleotide variants (SNVs) while evaluation of structural variants (SVs) remains most often performed by traditional cytogenetic approaches. Despite a rich history, cytogenetic studies - including chromosome banding analysis (CBA), Fluorescence In Situ Hybridization (FISH) and, more recently, chromosomal microarrays (CMA) - have well-described, method-specific diagnostic ‘blind spots’ which remain largely unaddressed (Table 1).5 There is thus a need to improve our capacity to robustly and reliably detect clinically relevant structural variants which may not be captured by more conventional methodologies.
Table 1. Comparison of Methodologies for Structural Variant Detection
Chromosome Banding Analysis (CBA) |
Fluorescent In Situ Hybridization (FISH) |
Chromosomal Microarray Analysis (CMA) |
Optical Genome Mapping (OGM) |
|
Coverage: |
Genome-Wide |
Targeted |
Genome-Wide |
Genome-Wide |
Specimen-Type |
Viable Cells |
Viable or Fixed Cells |
Viable or Fixed Cells |
Viable Cells(*) |
Analysis Type: |
Single Cell |
Single Cell |
Bulk Specimen |
Bulk Specimen |
Resolution: |
> 5 ~ 10 Mb |
~70 kb – 1 Mb |
> 5 kb ~ 200 kb |
> ~500 bp ~ 5 kb |
Sensitivity: |
~10%(**) |
~2-5% |
~10-15% |
5%-10% |
SVs types (***): |
CNV + SV |
CNV + SV |
CNV +/- AOH(****) |
CNV + SV + AOH |
(*) Viable cells can be frozen. Specifically designed DNA extraction protocol facilitates the isolation of ultra-high molecular weight (UHMW) DNA, 750ng of which is used each assay.
(**) While 20 metaphases are routinely reported for somatic-based testing, many more are often screened and thus the sensitivity may be lower than indicated
(***) For the purpose of this table, SV is delineated into copy number variants (CNVs) versus structural variations (SV) in which there is a change in the position of the genome.
(****) Only those chromosomal microarrays which incorporate SNPs permit the detection of region of absence or copy neutral loss of heterozygosity (AOH/CN-LOH),
Optical Genome Mapping (OGM) is an imaging technology which evaluates the fluorescent labeling pattern of individual DNA molecules to perform an unbiased assessment of genome-wide structural variants down to 500 base pairs (bp) in size,6 a resolution that far exceeds conventional cytogenetic approaches. OGM relies on a specifically designed extraction protocol facilitating the isolation of ultra-high molecular weight (UHMW) DNA. This protocol utilizes a paramagnetic disk purposed with trapping DNA for wash steps thereby reducing sheering forces present in standard column-based extraction methods. The result is DNA fragments of ~150 kilobases (kbp) to megabases (Mbp) in size, ~5-10x longer than the average fragment size from conventional DNA isolations techniques. DNA is fluorescently labeled via covalent modification at CTTAAG hexamer motifs, generating genome-wide density of approximately 14-17 labels per 100kb in sequence specific patterns.7 Labeled DNA is loaded on silicon chips composed of hundreds of thousands of parallel nanochannels where individual DNA molecules are linearized, imaged, and digitized. The specific labeling profile of individual DNA molecules – including spacing and pattern of hexamers labels – are subsequently grouped based on similarity, producing ~500 kb to megabase-sized consensus maps which can be compared in silico to the expected labeling pattern of a reference genome (Figure 1A). In essence, this imaging technology converts DNA into a “barcode” whose labeling profile and characteristics can sensitively and specifically resolve copy number and structural variation without the need for sequence level data (Figure 1A). The quality of the DNA – including both size and labeling characteristics – as well as the number of images captured influences genome-wide coverage. Presently, each flow cell, which can accommodate a single specimen - can generate up to 5000 Gigabase pairs (Gbp) of raw data, nearly a four-fold improvement from prior generations of this technology, achieving a maximum theoretical genome-wide coverage of ~1250x. Commercially developed informatic software supports several analyses including: de novo structural variant analysis for typical germline assessments (>~80x-coverage; requiring > ~400Gbp data collection) or ‘Rare Variant Analysis (RVP)’ for somatic assessment down to a ~5% variant allele fraction. (>~340x coverage; requiring >~1500 Gbp data). Both algorithms facilitate the detection of a wide array of structural variants; from copy number gains/losses to balanced/unbalanced translocations and insertions to inversions (Figure 1B). Notably, this assay can be completed in approximately ~4-days, the majority of which involves imaging and data analysis and thus requires limited hands-on technologist time (4~5 hours per run of 8~15 specimens).
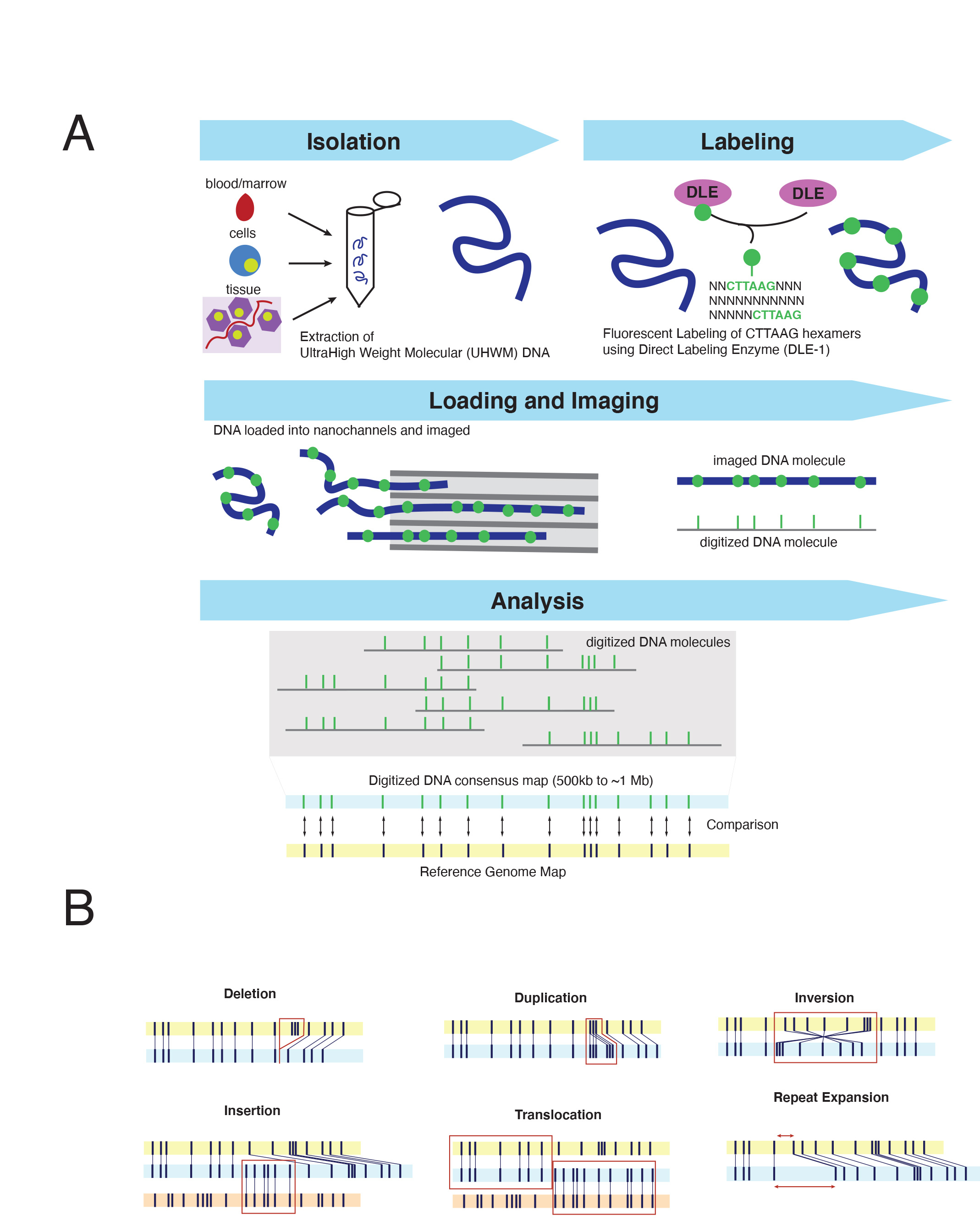
The end-to-end solution provided by OGM (from DNA isolation to data analysis) has facilitated a flurry of studies confirming the clinical diagnostic potential of this approach. While necessarily skewed towards hematologic malignancies, at the time of this publication over 1000 cases have been profiled by OGM, encompassing both myeloid and lymphoid neoplasms.8-13 These studies have collectively identified several recurrent themes. (1) OGM can reliably, and reproducibly detect all forms of clinically significant SVs that have been reported by standard of care testing Specifically, OGM has achieved >95% concordance with SVs detected by CBA, with discordances driven primarily by low-level whole chromosome aneuploidies (present in <5% of nuclei), which may be beyond the reliable detection threshold of this assay.14,15 (2) Present diagnostic standards for SV detection in hematologic malignancies fail to recognize clinically relevant aberrations at an alarming rate. In their study of MDS, Yang et al., noted that OGM identified clinically relevant SVs in 34% (34/100) of patients which were cytogenetically cryptic and therefore undetectable by CBA. Notably, in 17% (17/100) of their cohort, these findings would have changed the risk assessment, 15 a finding which was recapitulated in studies of AML in which OGM detected an additional 12-23% of SVs which would have altered clinical management or rendered patients eligible for clinical trials.14,16 While some of the inherent vulnerabilities of CBA could be supplemented by FISH-based studies (such as the detection of KMT2A and NUP98 rearrangements), OGM provides what both FISH and karyotype collectively can perform within a single assay.7 Similarly, in lymphoid cancers, OGM was able to assign major cytogenetic risk groups in 78% (32/41) of T and B-lineage precursor lymphoblastic leukemias, 20% greater than what can be accomplished by combined use of CBA, FISH and multiplex-ligation dependent probe amplification (MLPA).17 (3) Gene-level resolution of OGM improves understanding of cancer genomes and identifies clinically meaningful molecular subgroups. In 67% of myeloid malignancies studied, OGM analysis improved the resolution of breakpoints or clarified cytogenetic aberrations reported in by CBA. The enhanced resolution has led to expanding the capabilities of cytogenetic approaches to intragenic alterations, such as Partial Tandem Duplications (PTD) of KMT2A15, or PAX5.18
The promise of OGM for improved SV detection is not restricted to cancer-based testing. In germline-settings, where copy number variants (CNVs) detection is primarily performed by CMA, recent studies have shown that OGM has the capacity to detect all clinically relevant variants observed by standard of care studies.19 Moreover, OGM has been shown to effectively resolve complex chromosomal rearrangement (CCR) cases in the prenatal setting, assisting in characterizing the underlying genetic etiology of recurrent miscarriages.20 Moreover, germline studies have highlighted the potential for OGM to explore the ‘dark genome’ - repetitive regions of the genome which have been challenging to effectively characterize using Next Generation Sequencing (NGS) approaches. Repeat expansions - small (3-6 bp) nucleotide repeats which can be found tens to thousands of times in tandem - are associated with the onset of Fragile X syndrome, or myotonic dystrophy, among other conditions and can be successfully identified by OGM analyses if within the detection limits (i.e. > 500bp) of OGM analysis.21-23 Moreover, this approach led to the detection of novel mechanisms of pathogenesis, including the identification of a ~2.8 kb of a insertion within the intron 2 of SMARCB1 which was missed by whole genome sequencing due to the repetitive nature of the sequence coupled with the short-read nature of traditional NGS technologies, and provided a novel mechanism of inactivation leading to cancer predisposition.24
Despite these successes, it is important to note the inherent limitations of this technology which begins with the need for ultra-high weight molecular DNA. This precludes the capacity to evaluate specimens which have undergone fixation or to profile DNA that was isolated using conventional extractions. Moreover, not all specimens may yield effective isolation, which may be influenced by pre-analytical variables (specimen quality) or related to the technical performance of the isolation. OGM is also not presently a high-throughput technology. Each chip is composed of three flow-cells. At the time of this publication, current OGM workflows using a single instrument have capacity to analyze ~18-30 genomes per week. OGM also does not provide sequence level data and thus may require orthogonal, sequenced-based approaches to confirm certain classes of structural variants (i.e., small insertional events). Finally, with its increased detection of cryptic SVs, OGM may detect increased genomic variation of unknown significance and challenges current interpretative capabilities.
Taken together, while OGM shows the exquisite potential to significantly ameliorate the detection of clinically relevant structural variants above and beyond current cytogenetic approaches, its specific deployment as a possible clinical tool is best evaluated in the context of the totality of molecular and cytogenetic assays available at each institute. With the constant evolution in both understanding and technological capabilities, clinical diagnostic laboratories are presently faced with constraints regarding how best to prioritize the development of new assays. While SV-detection is beginning to undergo a radical transformation - from the resolution of gross chromosomal changes to nucleotide level solutions – the analytics and interpretation databases required to support nucleotide-level resolution is currently challenging for the typical clinical laboratory. As such, OGM represents a critical steppingstone to more finely resolved detection of structural lesions with relatively uniform and unbiased coverage of the genome.
References
- Ashley EA. Towards precision medicine. Nat Rev Genet. 2016;17(9):507-522. doi:10.1038/nrg.2016.86
- Levy-Sakin M, Ebenstein Y. Beyond sequencing: optical mapping of DNA in the age of nanotechnology and nanoscopy. Curr Opin Biotechnol. 2013;24(4):690-698. doi:10.1016/j.copbio.2013.01.009
- Berger MF, Van Allen EM. Delivering on the promise of precision cancer medicine. Genome Med. 2016;8(1):110. doi:10.1186/s13073-016-0373-1
- Friedman AA, Letai A, Fisher DE, Flaherty KT. Precision medicine for cancer with next-generation functional diagnostics. Nat Rev Cancer. 2015;15(12):747-756. doi:10.1038/nrc4015
- Akkari YMN, Baughn LB, Dubuc AM, et al. Guiding the global evolution of cytogenetic testing for hematologic malignancies. Blood. 2022;139(15):2273-2284. doi:10.1182/blood.2021014309
- Jeffet J, Margalit S, Michaeli Y, Ebenstein Y. Single-molecule optical genome mapping in nanochannels: multidisciplinarity at the nanoscale. Essays Biochem. 2021;65(1):51-66. doi:10.1042/EBC20200021
- Smith AC, Neveling K, Kanagal-Shamanna R. Optical genome mapping for structural variation analysis in hematologic malignancies. Am J Hematol. 2022;97(7):975-982. doi:10.1002/ajh.26587
- Nilius-Eliliwi V, Tembrink M, Gerding WM, et al. Broad genomic workup including optical genome mapping uncovers a DDX3X: MLLT10 gene fusion in acute myeloid leukemia. Front Oncol. 2022;12:959243. doi:10.3389/fonc.2022.959243
- Podvin B, Roynard P, Boudry A, et al. Whole-genome optical mapping to elucidate myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusions. Leuk Res. 2022;123:106972. doi:10.1016/j.leukres.2022.106972
- Puiggros A, Ramos-Campoy S, Kamaso J, et al. Optical genome mapping: a promising new tool to assess genomic complexity in chronic lymphocytic leukemia (CLL). Cancers (Basel). 2022;14(14). doi:10.3390/cancers14143376
- Ramos-Campoy S, Puiggros A, Kamaso J, et al. TP53 abnormalities are underlying the poor outcome associated with chromothripsis in chronic lymphocytic leukemia patients with complex karyotype. Cancers (Basel). 2022;14(15). doi:10.3390/cancers14153715
- Sahajpal NS, Mondal AK, Ananth S, et al. Clinical utility of combined optical genome mapping and 523-gene next generation sequencing panel for comprehensive evaluation of myeloid cancers. Published online January 17, 2022. doi:10.1101/2022.01.15.22269355
- Sahajpal NS, Mondal AK, Tvrdik T, et al. Clinical validation and diagnostic utility of optical genome mapping for enhanced cytogenomic analysis of hematological neoplasms. J Mol Diagn. 2022;24(12):1279-1291. doi:10.1016/j.jmoldx.2022.09.009
- Levy B, Baughn LB, Akkari Y, et al. Optical genome mapping in acute myeloid leukemia: a multicenter evaluation. Blood Adv. 2023;7(7):1297-1307. doi:10.1182/bloodadvances.2022007583
- Yang H, Garcia-Manero G, Sasaki K, et al. High-resolution structural variant profiling of myelodysplastic syndromes by optical genome mapping uncovers cryptic aberrations of prognostic and therapeutic significance. Leukemia. 2022;36(9):2306-2316. doi:10.1038/s41375-022-01652-8
- Gerding WM, Tembrink M, Nilius-Eliliwi V, et al. Optical genome mapping reveals additional prognostic information compared to conventional cytogenetics in AML/MDS patients. Int J Cancer. 2022;150(12):1998-2011. doi:10.1002/ijc.33942
- Rack K, De Bie J, Ameye G, et al. Optimizing the diagnostic workflow for acute lymphoblastic leukemia by optical genome mapping. Am J Hematol. 2022;97(5):548-561. doi:10.1002/ajh.26487
- Jean J, Kovach AE, Doan A, et al. Characterization of PAX5 intragenic tandem multiplication in pediatric B-lymphoblastic leukemia by optical genome mapping. Blood Adv. 2022;6(11):3343-3346. doi:10.1182/bloodadvances.2021006328
- Mantere T, Neveling K, Pebrel-Richard C, et al. Optical genome mapping enables constitutional chromosomal aberration detection. Am J Hum Genet. 2021;108(8):1409-1422. doi:10.1016/j.ajhg.2021.05.012
- Yang Y, Hao W. Identification of a familial complex chromosomal rearrangement by optical genome mapping. Mol Cytogenet. 2022;15(1):41. doi:10.1186/s13039-022-00619-9
- Morato Torres CA, Zafar F, Tsai YC, et al. ATTCT and ATTCC repeat expansions in the ATXN10 gene affect disease penetrance of spinocerebellar ataxia type 10. HGG Adv. 2022;3(4):100137. doi:10.1016/j.xhgg.2022.100137
- Otero BA, Poukalov K, Hildebrandt RP, et al. Transcriptome alterations in myotonic dystrophy frontal cortex. Cell Rep. 2021;34(3):108634. doi:10.1016/j.celrep.2020.108634
- Sahajpal NS, Barseghyan H, Kolhe R, Hastie A, Chaubey A. Optical genome mapping as a next-generation cytogenomic tool for detection of structural and copy number variations for prenatal genomic analyses. Genes (Basel). 2021;12(3). doi:10.3390/genes12030398
- Sabatella M, Mantere T, Waanders E, et al. Optical genome mapping identifies a germline retrotransposon insertion in SMARCB1 in two siblings with atypical teratoid rhabdoid tumors. J Pathol. 2021;255(2):202-211. doi:10.1002/path.5755

Samuel Brody, MS is a Technical Research Assistant II at Brigham and Woman Hospital. He is leading the effort to determine the diagnostic potential of Optical Genomic Mapping.

Adrian M. Dubuc, Ph.D FACMG is ABMGG dual certified Cytogeneticist and Molecular Geneticists who serves as an Assistant Laboratory Geneticists at Brigham and Women’s Hospital’s Center for Advanced Molecular Diagnostics (CAMD) and as Assistant Professor in Pathology at Harvard Medical School. Dr. Dubuc has served on numerous local and national committees contributing to the advancement of best practices in clinical laboratory genomic. This includes a tenure as the President of the Cancer Genomics Consortium (CGC) and both a chair and working member of numerous American College of Medical Genetics (ACMG) committees. In addition to his clinical role, Dr. Dubuc is a Principle Investigator whose research laboratory focuses on characterizing the structural and copy number landscape of cancer genomes. His work has led to over 80-peer reviewed publications, and resulted in the identifications of several clinically relevant molecular subgroups.

Annette S. Kim, MD, PhD, FCAP, is the Henry Clay Bryant Professor and Division Head of Diagnostic Genetics and Genomics at the University of Michigan. Dr. Kim’s research program has focused on the study of hematolymphoid malignancies, including miRNAs in myelodysplastic syndromes, myeloid and lymphoid mutational patterns, and test utilization management. She has served as a member of Molecular Oncology Committee and is currently chair of the Personized Healthcare Committee for College of American Pathologists. She is also program chair of the Association for Molecular Pathology and past chair of the ASH Subcommittee on Precision Medicine. In 2019, Dr. Kim was awarded the CAP Public Service Award, and in 2025 she received the CAP Laboratory Improvement Programs Service Award.